Our Research
Genome duplication during development
Housed within the nucleus of every cell in our bodies is six linear feet of DNA containing over three billion base pairs of DNA. Each time a cell divides, every single base pair of DNA must be accurately copied once and only once. To put the importance and scale of DNA replication in perspective, you will synthesize over one light year of DNA in your lifetime. The scale and complexity of DNA replication requires that hundreds of proteins function in a highly choreographed manner to ensure that our genomes are copied accurately and efficiently. A mistake copying just one single base of DNA can drive tumor formation or cell death. Furthermore, improper regulation of DNA replication has profound effects on human development. Meier-Gorlin syndrome (MGS) and Rothmund-Thomson syndrome (RTS), both characterized by several developmental abnormalities, are caused by mutations in key genes responsible for initiating DNA replication. Therefore, identification of factors involved in DNA replication could uncover causes of uncharacterized human disorders, aid in prenatal diagnosis and potentially permit prenatal therapies.
Much like studying the process of transcription in the context of development yielded the discovery of key transcriptional regulators, (e.g. Polycomb proteins), we study DNA replication in a developmental context to identify novel factors critical to replicate DNA and maintain genome stability using the fruit fly, Drosophila melanogaster, as a model organism. Our major focus is to identify proteins associated with the DNA replication machinery and delineate the underlying molecular mechanisms that these factors use to control genome duplication. Our ultimate goal is to understand how the process of DNA replication is regulated throughout the diversity of genome duplication programs that exist during development and identify how DNA replication becomes deregulated in disease states..
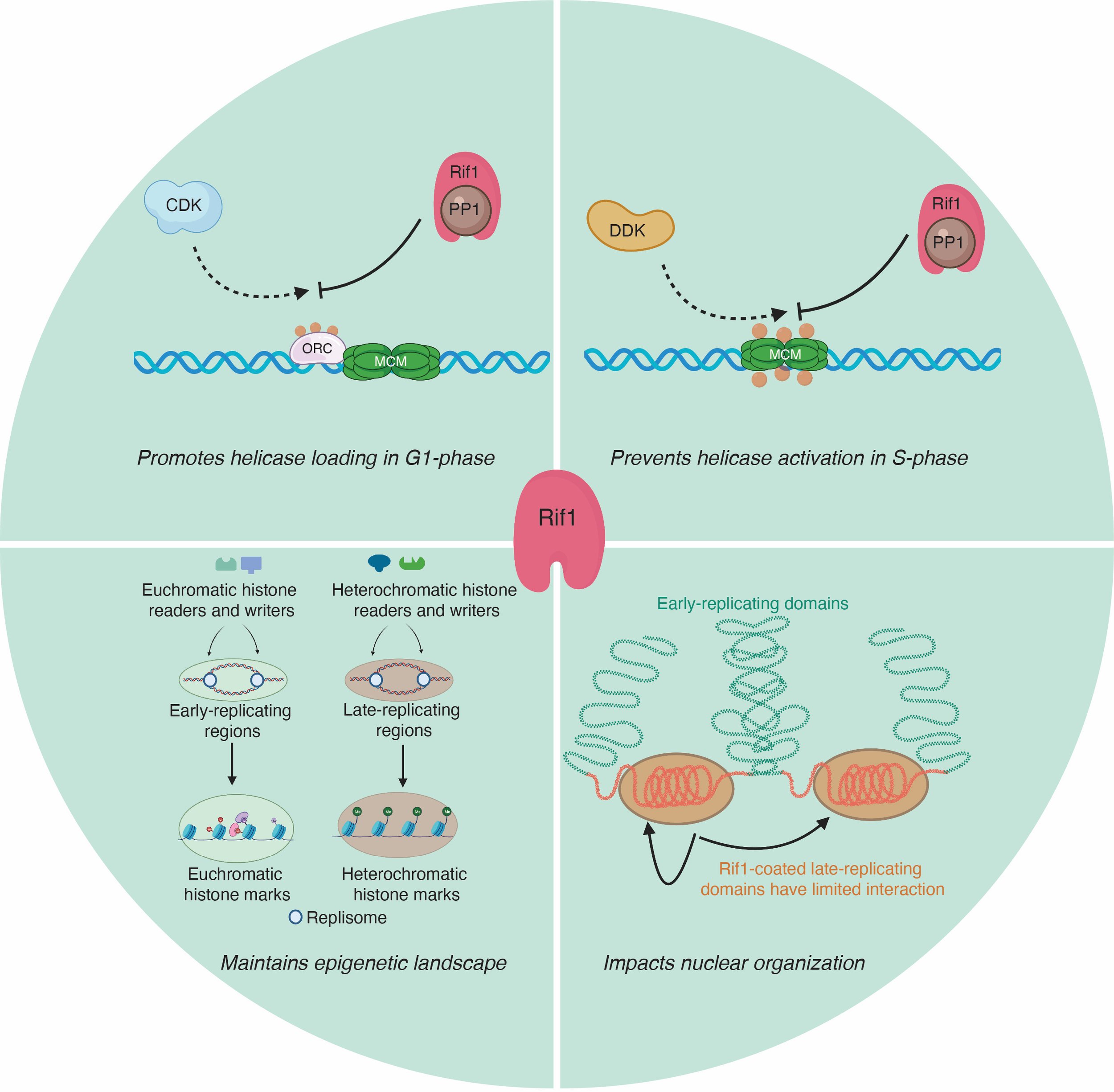
Rif1 control of replication during development
Our goal is to dissect mechanistically how Rif1 controls DNA replication using the powerful developmental systems of Drosophila. Work from multiple labs has revealed that Rif1 controls many aspects of DNA replication including; helicase activation, helicase loading and replication fork progression. The underlying molecular mechanisms that Rif1 uses to target and regulate key replication proteins remains unclear. We are using a combination of biochemical, proteomic, genetic, genomic and cell biological methods in the context of developoment to understand Rif1 function.
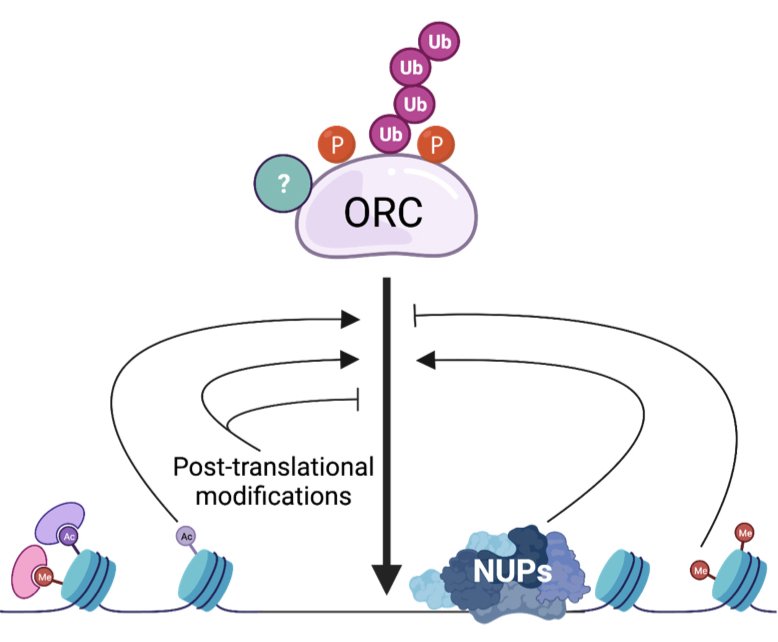
Regulation of ORC activity and localization
The Origin Recognition Complex (ORC) binds to thousands of sites throughout the genome to initiate DNA replication. The level of ORC binding throughout the genome is essential for efficient DNA replication and to maintain genome stability. In metazoans, ORC binding is sequence independent and highly influenced by chromatin. While we can map ORC-binding sites throughout metazoan genomes, understanding how ORC is loaded onto chromatin is still an outstanding question in the field. Equally important, is understanding how posttranslational modifications of ORC affect its ability to load the helicase throughout the genome. We are employing a diverse set of approaches to answer these questions.